The IIGM Italian Institute for Genomic Medicine participated in the WYMM Tour organized by Oxford Nanopore Technologies, which was recently held in Lyon.
During the event, was presented a retrospective study on selected cases of Breast and Ovarian Cancer, titled: “Implementation of Oxford Nanopore whole-genome sequencing for deciphering Large Genomic Rearrangements (LGRs) in familial breast cancer”.
It was a great opportunity to interact with researchers, clinicians, and the local and executive teams of Oxford Nanopore Technologies.
Furthermore, were discussed topics such as: Pharmacogenetics, Sequencing of Oncology Samples, and Single-Cell Approaches.
‘Implementation of Oxford Nanopore whole-genome sequencing for deciphering Large Genomic Rearrangements (LGRs) in familial breast cancer’
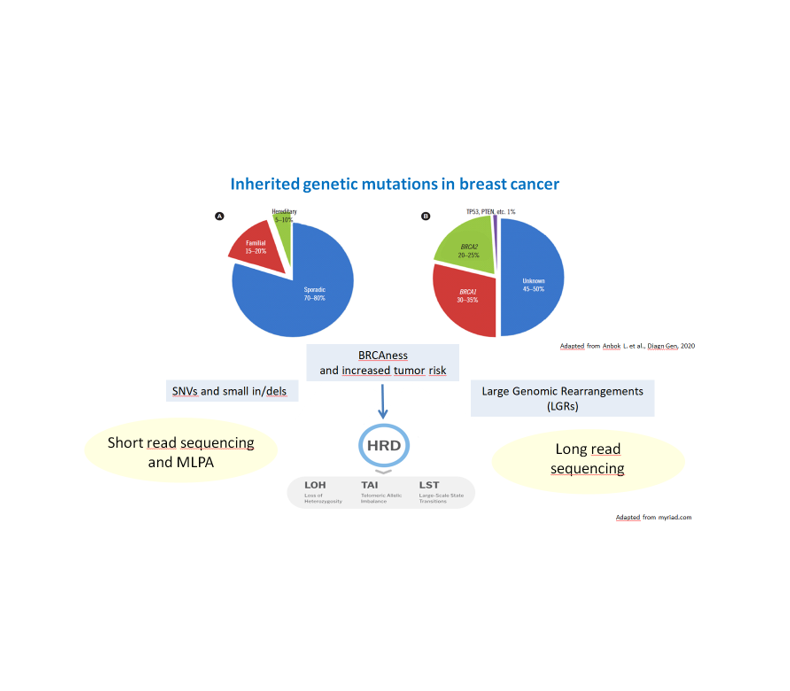
Background: Approximately 5–10% of breast cancers (BCs) are due to inherited genetic mutations. Among germline alterations, in addition to well-known pathogenic single nucleotide variations and small in/dels, large genomic rearrangements (LGRs) also contribute to the mutational landscape. It is therefore mandatory for clinical practice to develop fast and reliable diagnostic methods capable of resolving LGRs for patient management. Methods: We retrospectively analyzed a cohort of high-risk BC patients and their families (83 subjects). We extracted DNA from whole blood and prepared libraries for whole-genome sequencing (WGS) with the Ligation Sequencing DNA V14 kit following the manufacturer’s instructions, except for 5 samples that did not meet the protocol requirements, for which we implemented alternative strategies. We sequenced libraries on PromethION P24 and performed data QC using the Nanopore EPI2ME Human Variation workflow. Results: To ensure confident analysis, we aimed at achieving a sequencing depth of 30X (100 Gb) and an N50 greater than 5 kb. In the high-quality sample group, the success rate for depth and N50 was 96% and 98%, with a mean value of 123.6 Gb and 7.3 kb, respectively. In 17 out of 83 samples we needed to re-prepare or re-sequence the libraries. Thanks to optimization strategies we specifically devised, we achieved good results (93.4 Gb and 3.06 kb on average) also for the 5 suboptimal samples. Preliminary analysis of 22 samples confirmed that we achieved the target depth (37.11X on average). Conclusions: We have shown that ONT Ligation kit is suitable for performing long-read sequencing on whole-blood samples, even if stored frozen for a long time, allowing us to obtain data from 83 samples in less than 2 months. Overall, this approach needs optimization, but it could represent a potential valid and rapid alternative for diagnostic workflow for patients with familial breast cancer to resolve cases not associated with classical germline mutations.